25.2 Bootstrapping
How can we estimate the variability in a statistic? Let’s say we want a probability interval for the mean of some data.
One option is to rely on normal theory and write,
\[var(\bar{x}) = \frac{1}{n}var(x),\] then use the right normal score to get the ``confidence interval’’ you want:
xbar = mean(simData)
xsd = sd(simData)
# 95% interval
confInt = xbar + c(-1.96, 1.96) * (xsd/sqrt(n))
confInt
## [1] 1.800013 2.289007The accuracy of this estimate depends on the extent to which the data are normal. We know the data are from a rather skewed gamma, how much does that skewness affect the interval? How could we find out?
What if we had some large number of samples, say m, of the mean computed from \(n\) samples of this population? This is an awful idea in practice, but bear with me.
m = 2000
simulatedMeans = numeric(m)
# don't need for loop for this
for (i in 1:m) simulatedMeans[i] = mean(rgamma(n, 2, 1))We can visualize this:
plotSequence = seq(min(simulatedMeans), max(simulatedMeans), length = 1000)
hist(simulatedMeans, prob = TRUE, sub = "Line is the normal approximation density")
lines(plotSequence, dnorm(plotSequence, mean = xbar, sd = xsd/sqrt(n)))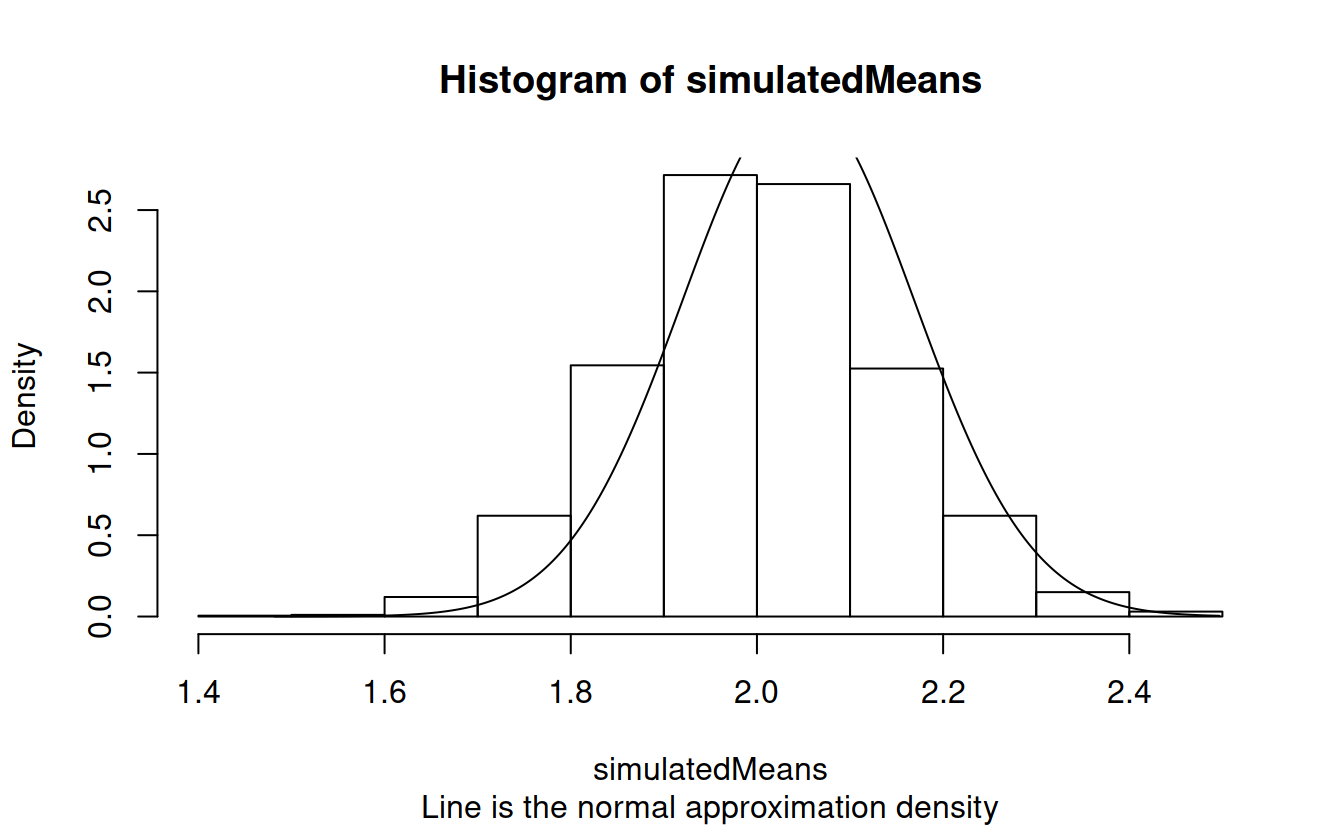
Obviously, we can’t just get a thousand means to estimate its variability, but we can do something close by resampling from our original data. This is called bootstrapping.
# b represents the number of bootstrap samples
b = 2000
bootstrapMeans = numeric(b)
for(i in 1:b){
resample = sample(simData, n, replace = TRUE)
bootstrapMeans[i] = mean(resample)
} hist(simulatedMeans, prob = TRUE, sub = "Line is the normal approximation density", col = rgb(1, 0, 0, .2))
hist(bootstrapMeans, prob = TRUE, col = rgb(0, 0, 1, .2), add = TRUE)
lines(plotSequence, dnorm(plotSequence, mean = xbar, sd = xsd/sqrt(n)))
legend("topright", bty = 'n', fill = c("red", "blue"), legend = c("True Mean Dist", "Bootstrap Dist"))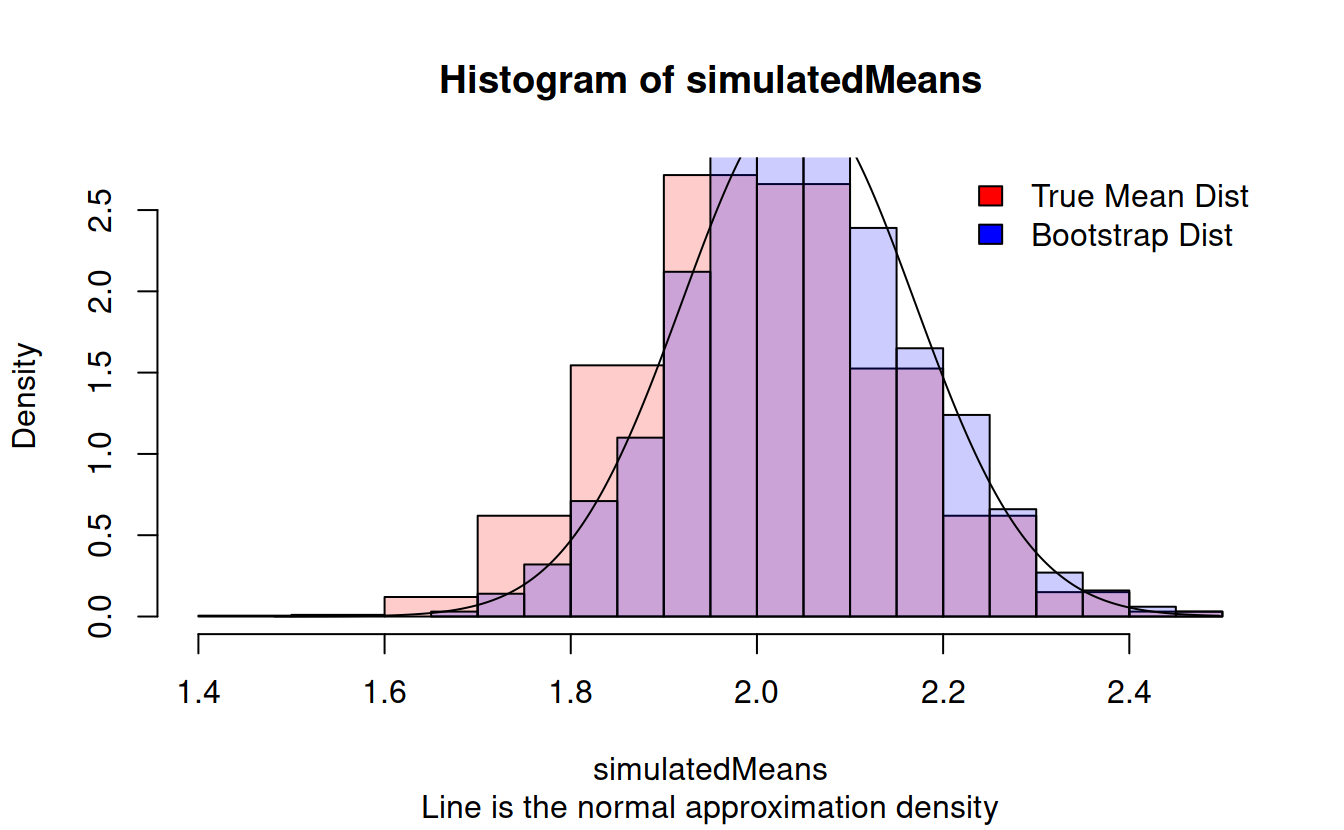
We see that this strange resampling technique seems to work well when we want to find a mean, at least as well as the normal approximation. Its true strength lies in estimating variability in more exotic statistics. For example, what is the variability in the width of the 95% confidence probability interval for our gamma data?
confintWidth = function(data){
upperAndLowerBounds = quantile(data, c(.025, .095))
width = diff(upperAndLowerBounds)
names(width) = NULL
return(width)
}m = 2000
simulatedWidths = numeric(m)
for (i in 1:m) simulatedWidths[i] = confintWidth(rgamma(n, 2, 1))
quantile(simulatedWidths, c(.025, .975))
## 2.5% 97.5%
## 0.1232144 0.4330531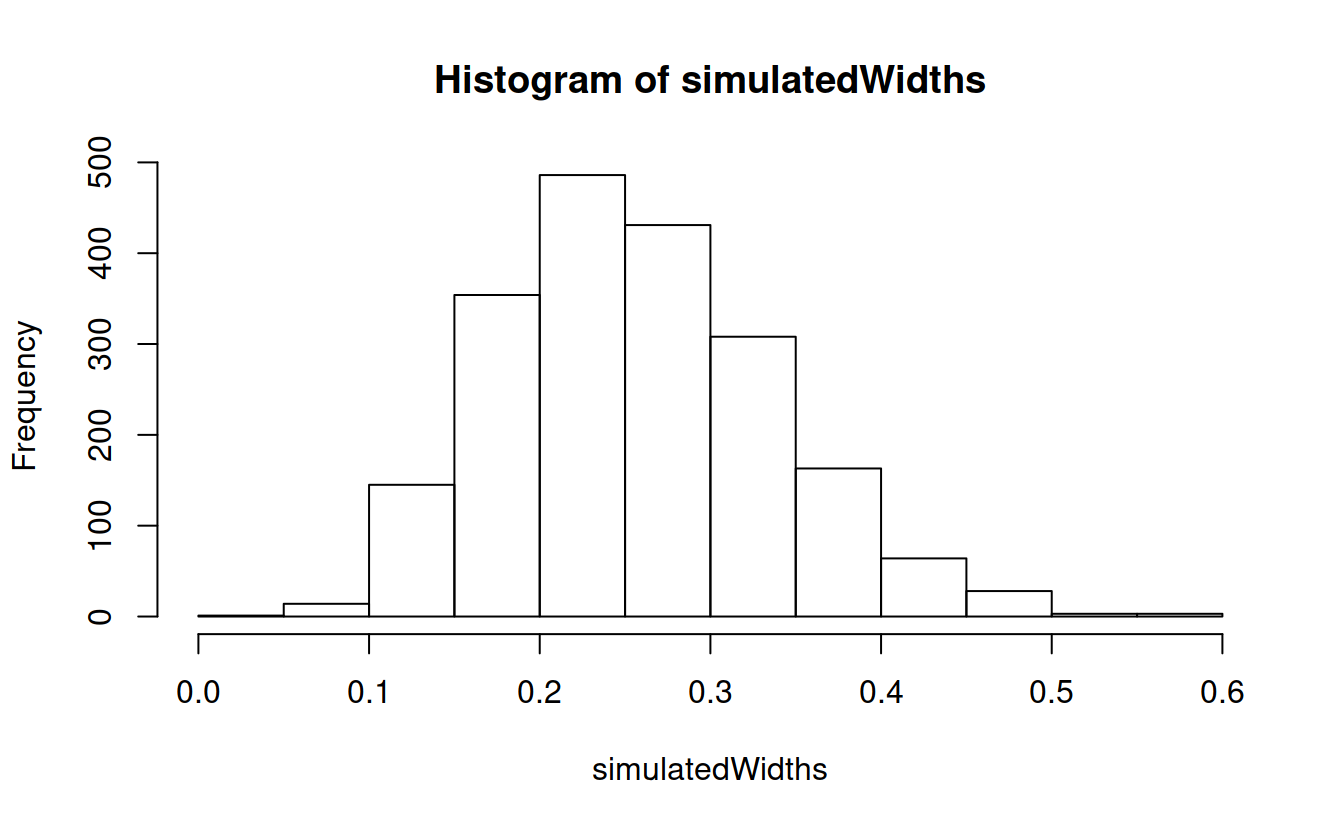
b = 2000
bootstrapWidths = numeric(b)
for(i in 1:b){
resample = sample(simData, n, replace = TRUE)
bootstrapWidths[i] = confintWidth(resample)
} hist(simulatedWidths, prob = TRUE, col = rgb(1, 0, 0, .2))
hist(bootstrapWidths, prob = TRUE, col = rgb(0, 0, 1, .2), add = TRUE)
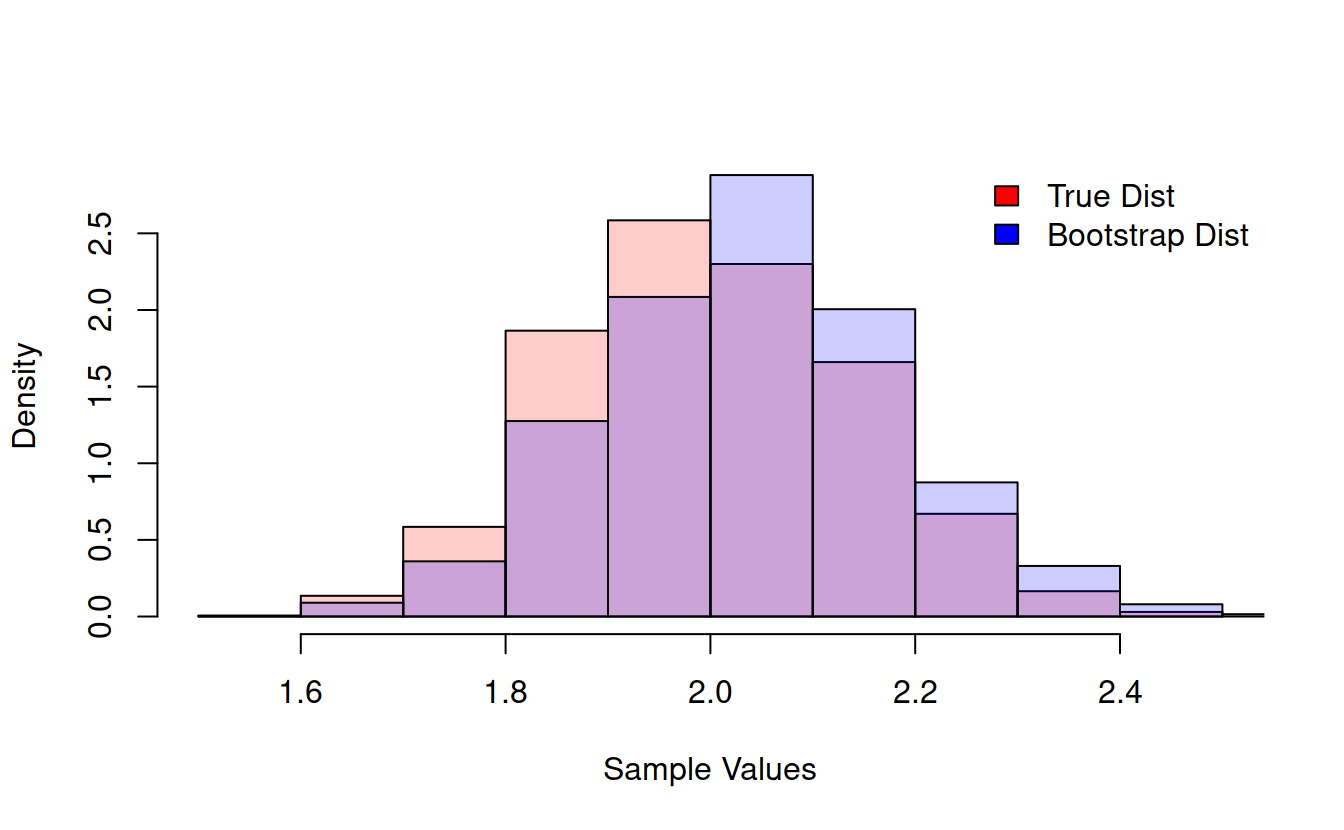
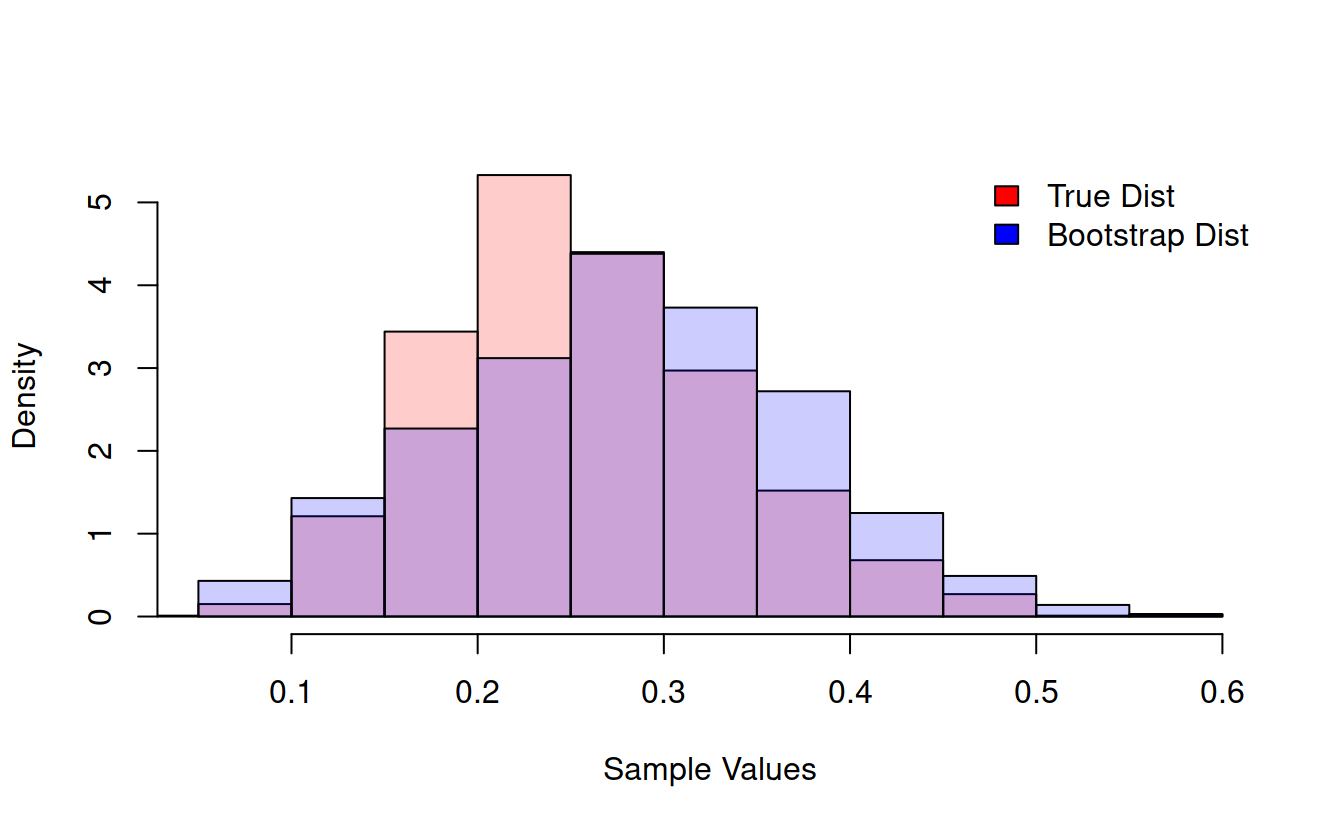
legend("topright", bty = 'n', fill = c("red", "blue"), legend = c("True Mean Dist", "Bootstrap Dist"))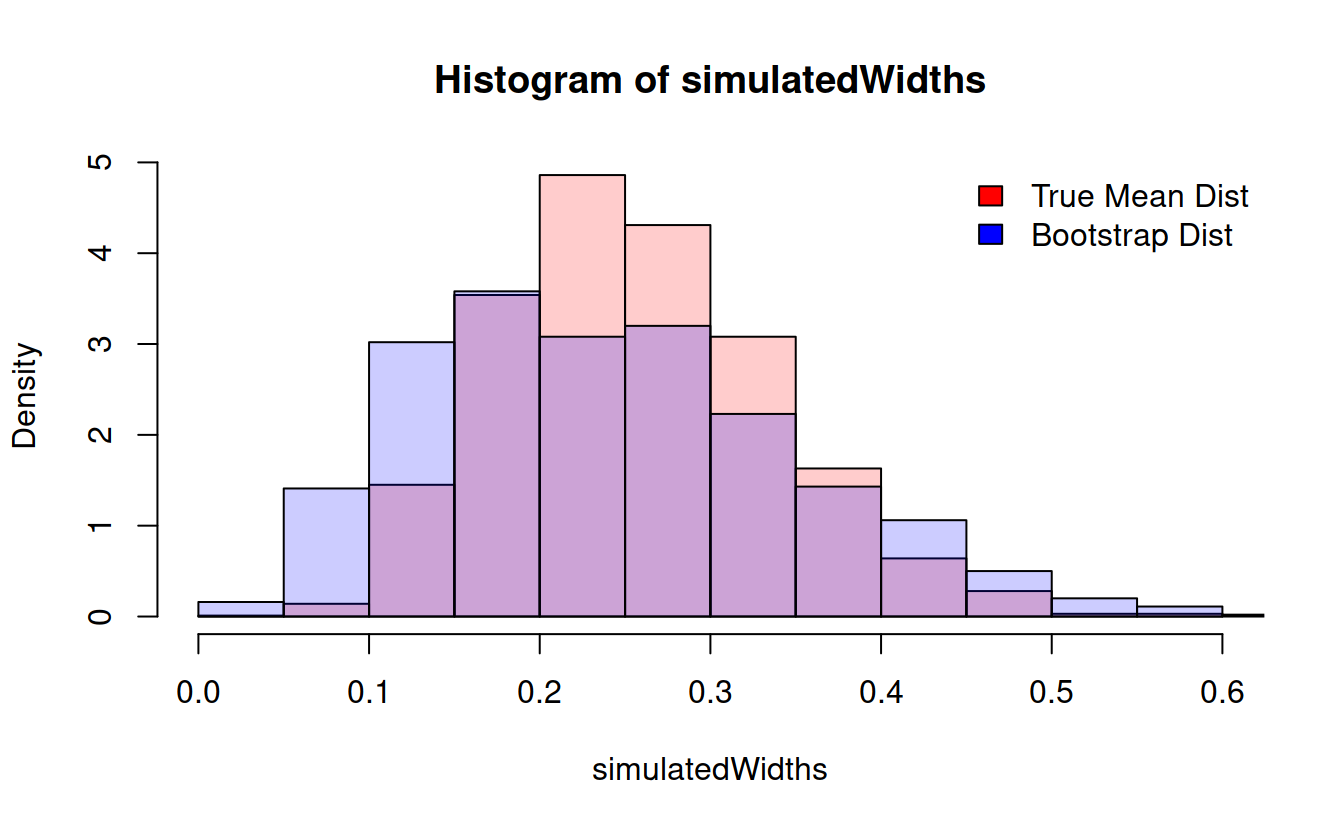 We can do this with any function at all!
We can do this with any function at all!
compareBootstrapToTruth = function(functionToCompare, sampleSize = 100, resamples = 1000, plot = FALSE, ret = FALSE){
data = rgamma(sampleSize, 2, 1)
trueSamples = numeric(resamples)
bootstrapSamples = numeric(resamples)
if(class(functionToCompare(data)) != "numeric") stop("Function must return a scalar.")
for(i in 1:m){
trueSamples[i] = functionToCompare(rgamma(sampleSize, 2, 1))
resample = sample(data, sampleSize, replace = TRUE)
bootstrapSamples[i] = functionToCompare(resample)
}
if(plot){
trueHist = hist(trueSamples, plot = FALSE)
bootHist = hist(bootstrapSamples, plot = FALSE)
hist(trueSamples, prob = TRUE, col = rgb(1, 0, 0, .2), main = "", xlab = "Sample Values", ylim = c(0, max(trueHist$density, bootHist$density)))
hist(bootstrapSamples, prob = TRUE, col = rgb(0, 0, 1, .2), add = TRUE)
legend("topright", bty = 'n', fill = c("red", "blue"), legend = c("True Dist", "Bootstrap Dist"))
}
out = list(trueSamples = trueSamples, bootstrapSamples = bootstrapSamples)
if(ret) return(out)
}